


|
|
Transformation:
introduction of recombinant DNA into an organism.
The earlier available techniques (biolistic, electroporation,
lithium-acetate or CaCl2/PEG techniques)
for transformation of fungi
require protoplasts or particle guns. Good protoplasts can be difficult to
produce consistently due to variations in enzyme batches (Ruiz-Die 2002).
In 1998 de Groot et al.
(de Groot et al. 1998),
successfully applied Agrobacterium tumefaciens mediated
transformation (also known as ATMT) to Aspergillus awamori, A.
niger, Fusarium venenatum, Trichoderma reesei, Colletotrichum
gloeosporioides, Neurospora crassa and the technique has later been
applied to F. oxysporum by Mullins et al. (2001). The technique had
up till then only been applied to dicot plants (since 1981 (Matzke & Chilton
1981)) and monocot plants (since 1993 (Chan et al. 1993)). The advantages is
that transformation can be carried out on conidia, spores or even vegetative
cells (Dobinson et al. 2004) (Mullins & Kang 2001). The technique showed a
600 fold increase in transformation efficiency for A. awamori
compared to protoplast-based techniques, and the majority of transformants
only harbors a single copy integration (de Groot et al. 1998) (Dobinson &
Kang 2001).
The gram-negative genus Agrobacterium comprises of both soil born
saprophytic and parasitic species. Many of the parasitic species cause
neoplastic diseases in plants, among others “hairy root disease” (A.
rhizogenes), “cane gall disease” (A. rubi), “crown gall of grape”
(A. vitis) and “crown gall disease/ crown gall tumors” (A.
tumefaciens) (Escobar & Dandekar 2003). They all use horizontal
interkindom gene transfer as a means to make the host plant provide suitable
conditions for survival of the bacteria.
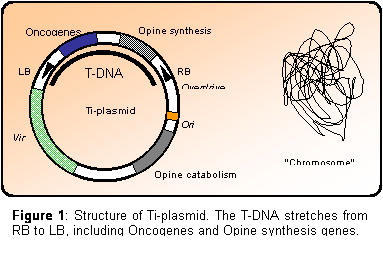
The genes located on the transferred DNA (termed T-DNA) can be divided into
two groups: The oncogenes, whose products (ultimately auxin and cytokinin)
cause the plant cells to proliferate, resulting in the formation of the
typical gall phenotype.
And the genes encoding opine like compounds, which
can be metabolized specifically by the infecting Agrobacterium, but
not by the plant itself. In this way, the bacteria induces the plant cells
to produce energy and nitrogen for the bacteria (the opine compound).
The T-DNA is located on a large Tumor inducing (Ti) plasmid (> 100 kb (Tkacz
et al. 2000)) which contains two additional classes of genes; Vir and
Opine catabolism. Vir genes encodes the proteins needed for
formation of T-DNA, T-pilus and transport of the T-DNA to the host cell
nucleus (Michielse et al. 2004). While the Opine catabolism genes
encodes the enzymes needed for the utilization of the opine compound
released by the plant host following successful transformation. The
boundaries of the T-DNA region is defined by two imperfect repeats (25 bp
long) called right and left border, RB and LB respectively (Figure
1).
Figure
1:
Structure of Ti-plasmid. The T-DNA stretches from RB to LB, including
Oncogenes and Opine synthesis genes.
|
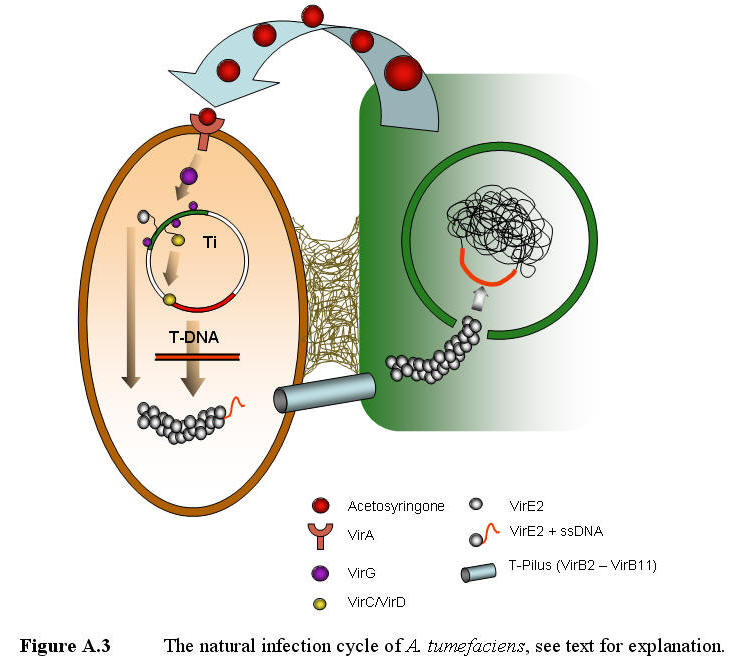
Agrobacterium
typically infect wounded plants. The release of plant saps, containing amino
acids, sugars and organic acids, attracts the bacteria to the wound by
positive chemotaxi. Once the bacteria reach the wound, it attaches itself to
the plant surface by synthesizing cellulose fibers (Escobar & Dandekar
2003). In addition to the mentioned metabolites wounded
plants also produce a wide range of phenolic compounds, such as coniferyl
alcohol and acetosyringone (AS) (Tkacz et al. 2000). These compounds induce
the bacteria to generate T-DNA, by a two component signaling system
(VirA/VirG). Acetosyringone activates VirA, a membrane bond receptor, which
activates the VirG (transcription factor). The activated VirG can then
interact with activator elements found in the promoters of the virA,
virB, virC, virD, virE and virG operons,
resulting in elevation of their expression levels (Figure A.3).
VirC and VirD (both nicking
endonucleases) binds to the RB/overdrive sequence and cuts the ssT-DNA
region out of the Ti plasmid. VirE2 binds to the ssT-DNA, protecting it from
degeneration by nucleases and self-annealing. VirB2-B11 forms a T-pilus
through which the VirE2 coated ssT-DNA is transferred from the bacteria into
the targeted plant cell (Zupan et al. 2003). The exact mechanism of this
process is not yet fully understood.
Inside the host cell, a
C-terminal located NLS in VirE2 directs the DNA into the nucleus, where host
factors are believed to facilitate its integration into the genome, possibly
mediated by the DNA repair system (Michielse et al 2004). If no great
sequence similarity exists between the plant genome and the introduced
T-DNA, the T-DNA integrates randomly into the nuclear genome
of the plant (Mullins
et al. 2001).
It was quickly realized that
any genetic material placed between the cis-acting RB and LB
sequences would be introduced into the genome of the targeted plant cells,
offering an easy way of producing transgenetic individuals (Matzke & Chilton
1981).
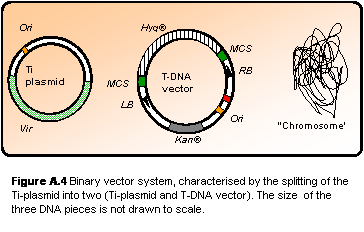
However, the large size of the
Ti-plasmid (> 100 kb) initially made it very difficult to handle and
manipulate by standard techniques. The solution was to move the T-DNA region
onto a smaller shuttle vector (Figure A.4). This is possible as the Vir
genes (still located on the originate Ti-plasmid) are trans-acting. Vector
systems of this type are called “binary vector systems”. These are also
disarmed as both the oncogenes and opine synthesis genes found in the wt
T-DNA has been replaced by selection marker genes (antibiotic resistance),
resulting in asymptomous infection. The opine catabolism genes on the
Ti-plasmid has also been removed, leaving only the Vir genes
necessary for the transformation machinery and structures that ensures
stable replication of the plasmid in A. tumefaciens (Hellens et al.
2000).
Many different T-DNA bearing
plasmids have been designed, but generally, three functional types can be
recognized (1) random mutagenesis, (2) promoter trapping and gene tagging
and (3) deletion of specific genes (Hellens et al. 2000).
|
Figure
A.4
Binary vector system, characterised by the splitting of the Ti-plasmid
into two (Ti-plasmid and T-DNA vector). The size of the three DNA
pieces is not drawn to scale.
|
The T-DNA normally integrates
randomly into the genome of the targeted cell. However if large segments of
the introduced DNA show a high degree of similarity to parts of the target
genome, this will “overwrite” the random integration, resulting in
homologous recombination. The length required for efficient homologous
recombination varies from specie to specie, but in general the success rate
is raised with increasing length of the segments showing similarity (Dobinson
et al. 2004).
References
 |
de Groot M.J.A., Bundock P.,
Hooykass P.J.J. and Beijersbergen “Agrobacterium tumefaciens-mediated
transformation of filamentous fungi”, Nature Biotechnology (1998), Vol. 16
p. 839-842 |
 |
Ruiz-Diez B. “Strategies for
the transformation of filamentous fungi” in Journal of Applied
Microbiology 2002, Vol. 92 p. 189-195 |
 |
Mullins E.D. and Kang S.
“Transformation: a tool for studying fungal pathogens of plants”, Cellular
and Molecular Life Sciences (2001), Vol. 58, p. 2043-3052. |
 |
Matzke A.J. and Chilton M.D.,
“Site-specific insertion of genes into T-DNA of the Agrobacterium
tumor-inducing plasmid: an approach to genetic engineering of higher plant
cells”, Journal of Molecular Applied Genetics (1981), Vol. 1, No. 1, p.
39-49 |
 |
Chan M.T., Chang H.H., Ho S.L.,
Tong W.F. and Yu S.M., “Agrobacterium-mediated production of
transgenic rice plants expressing a chimeric alpha-amylase promoter/beta-glucuronidase
gene”, Plant Molecular Biology (1993) Vol. 3, p. 491- 506 |
 |
Dobinson K.F., Grand S.J. and
Kang S. “Cloning and targeted disruption, via Agrobacterium
tumefaciens-mediated transformation, of a trypsin protease gene from
the vascular wilt fungus Verticillium dahliae”, Current
Genetics (2004), Vol. 45, p. 104-110 |
 |
Escobar M.A. and Dandekar A.M.
“Agrobacterium tumefaciens as an agent of disease”, TRENDS in Plant
Science 2003, Vol. 8 No. 8 p. 380-386 |
 |
Tkacz J.S., Dahl-Roshak A.M.,
Ibrahim N. and Paress P.S. “Handbook of Industrial Mycology” printed by
Marcel Dekker Inc , p. 3 (2000) |
 |
Michielse C.B., Ram A.F.,
Hooykaas P.J. and Hodel C.A. “Role of bacterial virulence proteins in
Agrobacterium-mediated transformation of Aspergillus awamori”,
Fungal Genetics and Biology (2004), Vol. 41, p. 571-578 |
 |
Zupan J., Muth T.R., Draper O.
and Sanbryski P. “The transfer of DNA from Agrobacterium tumefaciens into
plants: a feast of fundamental insights”, The Plant Journal (2003), Vol.
23 No. 1 p. 11-28 |
 |
Hellens R., Mullineax P. and
Klee H. “A guide to Agrobacterium binary Ti vectors”, TRENDS in
Plant Science (2000), Vol. 5 No. 10 p. 446- 451
|
|



